Abstract
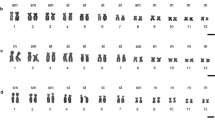
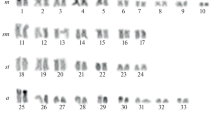
Chromosomal variation was analysed in 22 populations of newts of the salmandrid genusTaricha of western North America, and compared with that of the eastern North American newts of the genusNotophthalmus. The karyotypes of the speciesTaricha andNotophthalmus were very similar. However, there was considerable variation in the distribution patterns of heterochromatins (revealed by C-banding) and in the sites of the nucleolar organizing region (NOR) (revealed by fluorochrome chromomycin A3 banding) within and between species of these two genera. Chromosomal variation patterns were interpreted in relation to a phylogenetic hypothesis derived from data on mitochrondrial DNA sequences and allozyme variation. This study suggests that the pattern distributions of heterochromatins in chromosomes ofTaricha are more derived than those of its sister taxonNotophthalmus. Furthermore, the chromosomal NOR types found in the southernmost and northernmost populations ofT. granulosa, in the northernmost populations ofT. t. sierrae, and in the southern populations ofT. t. torosa are recently derived. The implications of this chromosomal variation for phylogeny and biogeography are discussed.
Similar content being viewed by others
References
Gall JG (1954) Lampbrush chromosomes from oocyte nuclei of the newt.J Morph 94: 283–352.
Green DM, Bogart JP, Anthony EH, Genner DL (1980) An interactive, microcomputer based karyotype analysis system for phylogenetic cytotaxonomy.Comput Biol Med 10: 219–227.
Hack MS, Lawce HJ (1980)The association of cytogenetic technologists: cytogenetics laboratory manual. San Francisco: University of California.
Haff T, Schmid M (1991) Immunocytogenetics: antinuclear antibodies as probes for chromosomal structure and function. In: Adolph KW, ed.Advanced Techniques in Chromosome Research. New York: Marcel Dekker, Inc. pp. 223–247.
Hayashi T, Matsui M (1989) Preliminary study of phylogeny in the family Salamandridae: allozyme data.Curr Herpetol E Asia 1989: 157–167.
Hedgecock D (1974)Protein variation and evolution in the genus Taricha (Salamandridae). Ph.D. dissertation, University of California, Davis.
Hsu TC (1981) Polymorphism in human acrocentric chromosomes and the silver staining method for nucleolus organizer regions.Karyogram 7: 45.
Hutchison N, Pardue ML (1975) The mitotic chromosomes ofNotophthalmus (= Triturus) viridescens: localization of C-banding regions and DNA sequences complementary to 18S, 28S and 5S ribosomal RNA.Chromosoma 53: 51–69.
Kezer J, Sessions SK (1979) Chromosome variation in the plethodontid salamanderAneides ferreus.Chromosoma 71: 65–80.
Léon PE (1976) Molecular hybridization of iodinated 4S, 5S, and 18+28S RNA to salamander chromosomes.J Cell Biol 69: 287–300.
Macgregor HC, Arntzen JW, Sessions SK (1990) An integrative analysis of phylogenetic relationships among newts of the genusTriturus (family Salamandridae), using comparative biochemistry, cytogenetics and reproductive interactions.J Evol Biol 3: 329–373.
Mancino G, Ragghianti M, Bucci-Innocenti S (1977) Cytotaxonomy and cytogenetics in European newt species. In: Tylor DH, Guttman SI, eds.The Reproductive Biology of Amphibians. New York: Plenum Press, pp. 411–447.
Morescalchi A (1973) Amphibia. In: Chiarelli AB, Capanna E, eds.Cytotaxonomy and Vertebrate Evolution. New York: Academic Press, pp. 233–348.
Morescalchi A. (1975) Chromosome evolution in the caudate amphibians.Evol Biol 8: 339–387.
Pukkila PJ (1975) Identification of the lampbrush chromosome loops which transcribe 5S ribosomal RNA inNotophthalmus (Triturus) viridescens.Chromosoma 53: 71–89.
Riemer WJ (1958) Variation and systematic relationships within the salamander genusTaricha.Univ California Publ Zool 56: 301–390.
Schmid M (1980) Chromosome banding in Amphibia. IV. Differentiation of GC- and AT-rich chromosome regions in Anura.Chromosoma 77: 83–103.
Schmid M, Guttenbach M (1988) Evolutionary diversity of reverse (R) fluorescent chromosome bands in vertebrates.Chromosoma 97: 101–114.
Schweizer D (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI.Chromosoma 58: 307–324.
Schweizer D, Loidl J, Hamilton B (1987) Heterochromatin and the phenomenon of chromosome banding. In: Hennig W, ed.Structure and Function of Eukaryotic Chromosomes. Berlin: Springer-Verlag, pp. 235–254.
Sessions SK, Kezer J (1987) Cytogenetic evolution in the plethodontid salamander genusAneides.Chromosoma 95: 17–30.
Seto T, Pomerat CM (1965)In vitro study of somatic chromosomes in newts, genusTaricha.Copeia 1965: 415–421.
Summer AT (1972) A simple technique for demonstrating centromeric heterochromatin.Exp Cell Res 75: 304–306.
Tan AM, Wu ZA, Zhao EM (1986a) Studies of the karyotype, C-bands and Ag-NORs ofRana liminocharis (Boie).Acta Herpetol Sinica 5: 176–180.
Tan AM, Wu ZA, Zhao EM, Ouyang HX (1986b) A handy one-step method for silver staining NORs.Acta Herpetol Sinica 5: 72–74.
Tan AM (1993)Systematics, phylogeny and biogeography of the northwest American newts of the genus Taricha (Caudata: Salamandridae). Ph.D. dissertation. University of California, Berkeley, pp. 296.
Tan AM, Wake DB (1993) Phylogeny and biogeography of the newt genusTaricha (Caudata, Salamandridae), inferred from mitochondrial cytochrome b DNA sequences. I. TheT. torosa species complex.Mol Phylogen and Evol, submitted.
Twitty, VC (1942) The species of CalifornianTriturus.Copeia 1942: 66–76.
Wake DB, Özeti N (1969) Evolutionary relationships in the family salamandridae.Copeia 1969: 124–137.
Wu ZA, Tan AM, Zhao EM (1987) Cytogenetic studies on four species ofAmolops in the Hengduan Range.Acta Gen Sinica 14: 63–68.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tan, AM. Chromosomal variation in the northwestern American newts of the genusTaricha (Caudata: Salamandridae). Chromosome Res 2, 281–292 (1994). https://doi.org/10.1007/BF01552722
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01552722