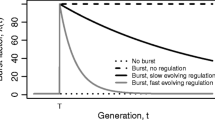
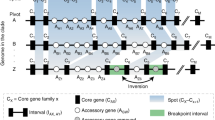
Abstract
A transposable element (TE) is a mobile sequence present in the genome of an organism. TEs can cause lethal mutations by inserting into essential, genes, promoting deletions or leaving short sequences upon excision. They therefore may be gradually eliminated from mixed populations of haploid micro-organisms such asEscherichia coli if they cannot balance this mutation load. Horizontal transmission between cells is known to occur and promote the transfer of TEs, but at rates often too low to compensate for the burden to their hosts. Therefore, alternative mechanisms should be found by these elements to earn their keep in the cells. Several theories have been suggested to explain their long-term maintenance in prokaryotic genomes, but little molecular evidence has been experimentally obtained. In this paper, the permanence of transposable elements in bacterial populations is discussed in terms of costs or benefits for the element and for the host. It is observed that, in all studies yet reported, the elements do not behave in their host as selfish DNA but as a co-operative component for the evolution of the couple.
Similar content being viewed by others
References
Arber., W., 1982. Das bacteriumE. coli under das Lupe der Molekulargenetiker. Mannheimer Forum 81/82. Herausgegeben von Boehringer Mannheim GmbH. 81p.
Arber, W., 1983. Bacterial inserted sequence elements and their influence on genetic stability and evolution. Proc. Nucleic Acids Res. 29:27–31.
Arber, W., 1990. Mechanisms in microbial evolution. J. Struct. Biol. 104:107–111.
Arber, W., 1991. Elements in microbial evolution. J. Mol. Evol. 33:4–12.
Arber, W., M. Hümbelin, P. Caspers, H.J. Reif, S. Iida & J. Meyer, 1980. Spontaneous mutations in theEscherichia coli prophage P1 and IS-mediated processes. Cold Spring Harbor Symp. Quant. Biol. 45:38–40.
Arber, W., T. Naas & M. Blot, 1994. Genetic rearrangements in resting bacteria. FEMS Microbiol. Ecol. (in press).
Belfort, M., 1990. Phage T4 introns: self-splicing and mobility. Ann. Rev. Genet. 24:363–385.
Berg, C.M., D.E. Berg & E.A. Grosman, 1989. Transposable elements and the genetic engineering of bacteria. pp 879–925 in Mobile DNA, edited by D.E. Berg and M. Howe. ASM, Washington D.C.
Biel, S.W. & D.L. Hartl., 1981. Beneficial effects of Tn5 are independent of transposition. Genetics 97:s11.
Biel, S.W. & D.L. Hartl, 1983. Evolution of transposons: natural selection for Tn5 inEscherichia coli K12. Genetics 103:581–592.
Blot, M., B. Hauer & G. Monnet, 1994. The Tn5-bleomycin resistance gene confers improved survival and growth advantage toEscherichia coli. Mol. Gen. Genet. 242:595–601.
Blot, M., J. Heitman & W. Arber, 1993. Tn5-mediated bleomycin resistance inEscherichia coli requires the expression, of host genes. Mol. Microbiol. 8:1017–1024.
Blot, M., J. Meyer & W. Arber, 1991. Bleomycin-resistance gene derived from the transposon Tn5 confers selective advantage toEscherichia coli K-12. Proc. Natl. Acad. Sci. USA. 88:9112–9116.
Campbell, A., 1981a. Evolutionary significance of accessory DNA elements in bacteria. Ann. Rev. Microbiol. 35:55–83.
Campbell, A., 1981b. Some questions about movable elemens and their implications. Cold Spring Harbor Symp. Quant. Biol. 45:1–9.
Campbell, A.M., D. Berg, D. Botstein, E. Lederberg, R. Novick, P. Starlinger & W. Szybalski, 1977. Nomenclature of transposable elements in prokaryotes. pp. 15–22 in DNA Insertion, Elements, Plasmids and Episomes, edited by A.I. Bukhari, J.A. Shapiro and S.L. Adhya. CSHL, Cold Spring Harbor.
Chao, L. & S.M. McBroom, 1985. Evolution of transposable elements: an IS10 insertion increases fitness inEscherichia coli. Mol. Biol. Evol. 2:359–369.
Chao, L., C. Vargas, B.B. Spear & E.C. Cox, 1983. Transposable elements as mutator genes in evolution. Nature 303:633–635.
Ciampi, M.S., M.B. Schmid & J.R. Roth, 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. USA 79:5016–5020.
Condit, R., 1990. The evolution of transposable elements: conditions for establishment in bacterial populations. Evolution 44:347–359.
Condit, R., F. Stewart & B. Levin, 1988. The population biology of bacterial transposons: a priori conditions for maintenance as parasitic DNA. Am. Nat. 132:129–147.
Datta, N., B.W. Randolph & J.L. Rosner, 1983. Detection of chemicals that stimulate Tn9 tranposition inEscherichia coli. Mol. Gen. Genet. 189:245–250.
Doolittle, W.F. & C. Sapienza, 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603.
Edlin, G., S.W. Lee & M.M. Green, 1986. Tn10 transposition does not respond to environmental stress. Mut. Res. 175:159–164.
Escoubas, J.M., M.F. Prère, O. Fayet, I. Salvignol, D. Galas, D. Zerbib & M. Chandler, 1991. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 10:705–712.
Galas, D.J. & M. Chandler, 1989. Bacterial Insertion Sequences, pp. 109–162 in Mocile DNA, edited by D. Berg and M. Howe. ASM, Washington D.C.
Hall, B.G., 1988. Adaptive evolution that requires multiple spontaneous mutations I. Mutations involving an insertion sequence. Genetics 120:887–897.
Hall, B.G., 1991. Is the occurrence of some spontaneous mutations directed by environmental challenges? The New Biologist 3:729–733.
Hartl, D.L., D.E. Dykhuizen, R.D. Miller, L. Green & J. De Framond, 1983. Transposable element IS50 improves growth rate ofEscherichia coli cells without transposition. Cell 35:503–510.
Inouye, M. & J.S. Inouye, 1992. Retrons and multicopy single stranded DNA. J. Bacteriol. 174:2419–2424.
Jilk, R.A., J.C. Makris, L. Borchardt & W.S. Reznikoff, 1993. Implications of Tn5-Associated adjacent deletions. J. Bacteriol. 175:1264–1271.
Kolter, R., D.A. Siegele & A. Tormo, 1993. The stationary phase of the bacterial cycle. Ann. Rev. Microbiol. 47:855–74.
Kurlandzka, A., R.F. Rosenzweig & A. Adams, 1991. Identification of adaptive changes in an evolving population ofEscherichia coli: the role of changes with regulatory and highly pleiotropic effects. Mol. Biol. Evol. 8:261–281.
Lawrence, J.G., H. Ochman & D.L. Hartl, 1992. The evolution of insertion sequences within enteric bacteria. Genetics 131:9–20.
Médigue, C., T. Roxel, P. Vigier, A. Hénaut & A. Danchin, 1991. Evidence for horizontal gene transfer inEscherichia coli speciation. J. Mol. Biol. 222:851–856.
Mikkola, R. & C.G. Kurland, 1991. Is there a unique ribosome phenotype for naturally occurringEscherichia coli? Biochimie 73:1061–1066.
Mikkola, R. & C.G. Kurland, 1992. Selection of laboratory wild-type phenotype from natural isolates ofEscherichia coli in chemostats. Mol. Biol. Evol. 9:394–402.
Modi, R.I., L.H. Castilla, S. Puskas-Rozsa, R.B. Helling & J. Adams, 1992. Genetic changes accompanying increased fitness in evolving populations ofEscherichia coli. Genetics 130:241–249.
Naas, T., M. Blot, W.M. Fitch & W. Arber, 1994. Insertion sequence-related genetic rearrangements in restingEscherichia coli K-12. Genetics 136:721–730.
Orgel, L.E. & F.H.C. Crick, 1980. Selfish DNA: the ultimate parasite. Nature 284:604–607.
Plasterk, R.H.A., 1991. Frameshift control of IS1 transposition. Trends Genet. 7:203–204.
Raabe, T., E. Jenny & J. Meyer, 1988. A selection cartridge for rapid detection and analysis of spontaneous mutations including insertions of transposable elements in Enterobacteriaceae. Mol. Gen. Genet. 215:176–180.
Rodriguez, H., E.T. Snow, U. Bhat & E.L. Loechler, 1992. AnEscherichia coli plasmid-based, mutational system in whichsupF mutants are selectable — insertion elements dominate the spontaneous spectra. Mut. Res. 270:219–231.
Ross, D.G., J. Swan & N. Kleckner, 1979. Nearly precise excision: a new type of DNA alteration associated, with the translocatable element Tn10. Cell 16:733–738.
Rusina, O.Y., E.E. Mirskaya, I.V. Andreeva & A.G. Skavronskaya, 1992. Precise excision of transposons and point mutations induced by chemicals. Mut. Res. 283:161–168.
Sawyer, S.A., D.E. Dykhuizen, R.F. DuBose, L. Green, T. Mutangadure-Mhlanga, D.F. Wolczyk & D.L. Hartl, 1987. Distribution and abundance of insertion sequences among natural isolates ofEscherichia coli. Genetics 115:51–63.
Schnetz, K. & B. Rak, 1992. IS5 — A mobile enhancer of transcription inEscherichia coli. Proc. Natl. Acad. Sci. USA. 89:1244–1248.
Scott, J.R., 1992. Sex and the single circle: conjugative transposition. J. Bacteriol. 174:6005–6018.
Sekine, Y. & E. Ohtsubo, 1989. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc. Natl. Acad. Sci. USA, 86:4609–4613.
Sengstag, C. & W. Arber, 1983. IS2 insertion, is a major cause of spontaneous mutagenesis of the bacteriophage P1: non random distribution of target sites. EMBO J. 2:67–71.
Toussaint, A. & A. Résibois, 1983. Phage Mu: transposition as a life-style, pp. 105–158 in Mobile Genetic Elements edited by J. Shapiro, Acad. Press. Inc., Orlando.
Wang, A. & J.R. Roth, 1988. Activation of silent genes by transposons Tn5 and Tn10. Genetics 120:875–885.
Young, J.P.W., 1989. The population genetics of bacteria. pp 417–438 in Genetics of Bacterial Diversity edited by D.A. Hopwood and K.E. Chater, Acad. Press Lim, London.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Blot, M. Transposable elements and adaptation of host bacteria. Genetica 93, 5–12 (1994). https://doi.org/10.1007/BF01435235
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01435235