Abstract
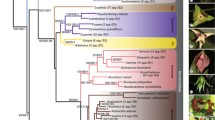
A molecular phylogeny of the familyIridaceae based on the plastid generps4 was obtained using both parsimony and distance methods. Thirty-four species were examined together with eight outgroup species. Results show that theIridaceae are monophyletic, and thatIsophysis is likely to be the earliest emerging genus. SubfamilyIxioideae plus the generaAristea andNivenia form a strongly supported clade. Within subfam.Iridoideae, the tribeIrideae includes the genusBobartia (of disputed position), and the tribeMariceae includesCypella. The division ofIridoideae into tribes is consistent with their geographical distribution.
Similar content being viewed by others
References
Bittar, G., 1995: Phylogénie: histoire, concepts, principes et présentation de nouvelles méthodes d'analyse numérique de l'évolution moléculaire. — Ph.D. Dissertation, Université de Genève, Switzerland.
-Carter, L., 1994: New probabilistic and numerical phenetics methods for inferring natural groupings and phylogenesis. — In: Proceedings of the International Meeting “Ecology and statistical methods”, Niort, France, pp. 145–150.
Bremer, K., 1988: The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. — Evolution42: 795–803.
Chase, M. W., Soltis, D. W., Olmstead, R. G., Morgan, D., Les, D. H., Mishler, B. D., Duvall, M. R., Price, R. A., Hills, H. G., Qiu, Y. L., Kron, K. A., Retting, J. H., Conti, E., Palmer, J. D., Manhart, J. R., Systma, K. J., Michaels, H. J., Kress, W. J., Karol, K. G., Clark, W. D., Hedren, M., Gaut, B. S., Jansen, R. K., Kim, K. J., Wimpee, C. F., Smith, J. F., Furnier, G. R., Strauss, S. H., Xiang, Q. Y., Plunkett, G. M., Soltis, P. S., Swensen, S. M., Williams, S. E., Gadek, P. A., Quinn, C. J., Eguiarte, L. E., Golenberg, E., Learn, G. H. J., Graham, S. W., Barrett, S. C. H., Dayanandan, S., Albert, V. A., 1993: Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid generbcL. — Ann. Missouri Bot. Gard.80: 528–580.
, 1995: Molecular phylogenetics ofLilianae. — InRudall, P. J., Cribb, P. J., Cutler, D. F., Humphries, C. J., (Eds): Monocotyledons: systematics and evolution,1: pp. 109–137. — Kew: Royal Botanic Gardens.
Clegg, M. T., Zurawski, G., 1992: Chloroplast DNA and the study of plant phylogeny: present status and future prospects. — InSoltis, P. S., Soltis, D. E., Doyle, J. J., (Eds): Molecular systematics of plants, pp. 1–13. — New York: Chapman & Hall.
, 1994: Rates and patterns of chloroplast DNA evolution. — Proc. Natl. Acad. Sci. USA91: 6795–6801.
Cronquist, A., 1981: An integrated system of classification of flowering plants. — New York: Columbia University Press.
Dahlgren, R. M. T., Clifford, H. T., Yeo, P. F., 1985: The families of monocotyledons. — Berlin, Heidelberg, New York: Springer.
Doyle, J. J., Doyle, J. L., 1987: A rapid DNA isolation procedure for small quantities of fresh leaf tissue. — Phytochem. Bull.19: 11–15.
Duvall, M. R., Clegg, M. T., Chase, M. W., Clark, W. D., Kress, W. J., Hills, H. G., Eguiarte, L. E., Smith, J. F., Gaut, B. S., Zimmer, E. A., Learn, G. H., 1993: Phylogenetic hypothesis for the monocotyledons constructed fromrbcL sequence data. — Ann. Missouri Bot. Gard.80: 607–619.
Felsenstein, J., 1985: Confidence limits on phylogenies: an approach using the bootstrap. — Evolution39: 783–791.
Gielly, L., Taberlet, P., 1994: The use of chloroplast DNA to resolve plant phylogenies: non-coding versusrbcL sequences. — Molec. Biol. Evol.11: 769–777.
Goldblatt, P., 1990: Phylogeny and classification ofIridaceae. — Ann. Missouri Bot. Gard.77: 607–627.
, 1991: An overview of the systematics, phylogeny and biology of the AfricanIridaceae. — Contr. Bolus Herb.13: 1–74.
, 1992: Relationships ofBobartia. — S. African J. Bot.58: 304–309.
, 1995: Pollination biology ofLapeirousia subgenusLapeirousia (Iridaceae) in southern Africa; floral divergence and adaptation for longtongued fly pollination. — Ann. Missouri Bot. Gard.82: 517–534.
Ham, R. C. H. J. v., Hart, H., Mes, T. H. M., Sandbrink, J. M., 1994: Molecular evolution of non-coding regions of the chloroplast genome in theCrassulaceae and related species. — Curr. Genet.25: 558–566.
Hoot, S. B., Culham, A., Crane, P. R., 1995: The utility ofatpB gene sequences in resolving phylogenetic relationships: comparison withrbcL and 18S ribosomal DNA sequences in theLardizabalaceae. — Ann. Missouri Bot. Gard.82: 194–207.
Johnson, L. A., Soltis, D. E., 1994:matK DNA sequences and phylogenetic reconstruction inSaxifragaceae s.str. — Syst. Bot.19: 143–156.
,, 1995: Phylogenetic inference inSaxifragaceae sensu stricto andGilia (Polemoniaceae) usingmatK sequences. — Ann. Missouri Bot. Gard.82: 149–175.
Lamppa, G. K., Bendich, A. J., 1979: Chloroplast DNA sequence homologies among vascular plants. — Pl. Physiol.63: 660–668.
Lavin, M., Doyle, J. J., Palmer, J. D., 1990: Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in theLeguminosae subfam.Papilionoideae. — Evolution44: 390–402.
Maddison, W. P., Maddison, D. R., 1992: MacClade: analysis of phylogeny and character evolution. Version 3.0. — Sunderland, Mass.: Sinauer.
Manen, J. F., Natali, A., Ehrendorfer, F., 1994: Phylogeny ofRubiaceae-Rubieae inferred from the sequence of a cpDNA intergene region. — Pl. Syst. Evol.190: 195–211.
Mes, T. H. M., Hart, H., 1994:Sedum surculosum andS. jaccardianum (Crassulaceae) share a unique 70 bp deletion in the chloroplast DNAtrnL (UAA)—trnF (GAA) intergenic spacer. — Pl. Syst. Evol.193: 213–221.
Nadot, S., 1994: Utilisation de l'outil moléculaire pour la reconstruction d'une phylogénie des plantes monocotylédones en général et de la famille desPoaceae (Graminées) en particulier. — Ph.D. in Sciences — Dissertation, Université de Paris XI, Orsay, France.
, Bajon, R., Lejeune, B., 1994: The chloroplast generps 4 as a tool for the study ofPoaceae phylogeny. — Pl. Syst. Evol.191: 27–38.
, Bittar, G., Carter, L., Lacroix, R., Lejeune, B., 1995: A phylogenetic analysis of monocotyledons based on the chloroplast generps 4, using parsimony and a new numerical phenetics method. — Molec. Phylogenet. Evol.4: 257–282.
Olmstead, R. G., Reeves, P. A., 1995: Evidence for the polyphyly of theScrophulariaceae based on chloroplastrbcL andndhF sequences. — Ann. Missouri Bot. Gard.82: 176–193.
Palmer, J. D., Zamir, D., 1982: Chloroplast DNA evolution and phylogenetic relationships inLycopersicum. — Proc. Natl. Acad. Sci. USA79: 5006–5010.
, Stein, D. B., 1986: Conservation of chloroplast genome structure among vascular plants. — Curr. Genet.10: 823–833.
Ritland, K., Clegg, M. T., 1987: Evolutionary analysis of plant DNA sequences. — Amer. Naturalist130: S74-S100.
Rudall, P., 1994: Anatomy and systematics ofIridaceae. — Bot. J. Linn. Soc.114: 1–21.
Soltis, D. E., Soltis, P. S., Ness, B. D., 1989: Chloroplast-DNA variation and multiple origins of autopolyploidy inHeuchera micrantha (Saxifragaceae). — Evolution43: 650–656.
Steele, K. P., Vilgalys, R., 1994: Phylogenetic analyses ofPolemoniaceae using nucleotide sequences of the plastid genematk. — Syst. Bot.19: 126–142.
Swofford, D. L., 1993: PAUP 3.1: phylogenetic analysis using parsimony. — Washington, D.C.: Smithsonian Institution Press.
Taberlet, P., Gielly, L., Pautou, G., Bouvet, J., 1991: Universal primers for amplification of three non-coding regions of chloroplast DNA. — Pl. Molec. Biol.17: 1105–1109.
Thompson, J. D., Higgins, D. G., Gibson, T. J., 1994: Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. — Nucl. Acids Res.22: 4673–4680.
Wolfe, K. H., Morden, C. W., Palmer, J. D., 1991: Ins and outs of plastid genome evolution. — Curr. Opinion Genet. Developm.1: 523–529.
,, Ems, S. C., Palmer, J. D., 1992: Rapid evolution of the plastid translational apparatus in a non-photosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. — J. Molec. Evol.35: 304–317.
Zurawski, G., Clegg, M. T., 1987: Evolution of higher-plant chloroplast DNA-encoded genes: implications for structure-function and phylogenetic studies. — Annu. Rev. Pl. Physiol.38: 391–418.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Souza-Chies, T.T., Bittar, G., Nadot, S. et al. Phylogenetic analysis ofIridaceae with parsimony and distance methods using the plastid generps4. Pl Syst Evol 204, 109–123 (1997). https://doi.org/10.1007/BF00982535
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00982535