Summary
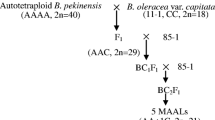
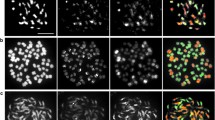
This study aimed at generating chromosome addition lines and disclosing genome specific markers in Brassica. These stocks will be used to study genome evolution in Brassica oleracea L., B. campestris L. and the derived amphidiploid species B. napus L. B. campestris-oleracea monosomic and disomic chromosome addition plants were generated by crossing and backcrossing the natural amphidiploid B. napus to the diploid parental species B. campestris. The pollen viability of the derived sesquidiploid and hyperploid ranged from 63% to 88%, while the monosomic and disomic addition plants had an average pollen fertility of 94% and 91%, respectively. The addition lines were genetically characterized by genome specific markers. The isozymes for 6PGD, LAP, PGI and PGM, and rDNA Eco RI restriction fragments were found to possess the desired genome specificity. Duplicated loci for several of these markers were observed in B. campestris and B. oleracea, supporting the hypothesis that these diploid species are actually secondary polyploids. A total of eight monosomic and eight disomic addition plants were identified and characterized on the basis of these markers. Another 51 plants remained uncharacterized due to the lack of additional markers. rDNA genes were found to be distributed in more than one chromosome, differing in its restriction sites. Intergenomic recombination for some of the markers was detected at frequencies between 6% and 20%, revealing the feasibility of intergenomic gene transfer.
Similar content being viewed by others
References
Appels R, Dvorak J (1982) Relative rates of divergence of spacer and gene sequences within the rDNA region of species in the Triticeae: implications for maintenance of homogeneity of the repeated gene family. Theor Appl Genet 63:361–365
Arus P (1984) B. oleracea and B. napus isozymes. Cruciferae Newslett 9:64
Arus P, Orton TJ (1983) Isozyme and linkage relationships of isozyme loci in Brassica oleracea. J Hered 74:405–412
Attia T, Robbelen G (1986) Cytogenetic relationship within cultivated Brassica analyzed in amphihaploids from three diploid ancestors. Can J Genet Cytol 28:323–329
Catcheside DG (1934) The chromosomal relationships in the swede and turnip groups of Brassica. Ann Bot 48:601–633
Catcheside DG (1937) Secondary pairing in Brassica oleracea Cytologia. Jubilee, pp 366–378
Chiang MS, Crete R (1983) Transfer of resistance to race 2 of Plasmodiophora brassicae from Brassica napus to cabbage (B. oleracea ssp. capitata). V. The inheritance of resistance. Euphytica 32:479–483
Coulthart M, Denford KE (1982) Isozyme studies in Brassica. I. Electrophoretic techniques for leaf enzymes and comparison of B. napus, B. compestris and B. oleracea using phosphoglucomutase. Can J Plant Sci 62:621–630
Delseny M, Cooke R, Penon P (1983) Sequence heterogeneity in radish nuclear ribosomal RNA genes. Plant Sci Lett 30:107–109
Erickson LR, Straus NA, Beversdorf WD (1983) Restriction patterns reveal origins of chloroplast genomes in Brassica amphidiploids. Theor Appl Genet 65:201–206
Hart GE, Tuleen NA (1983) Chromosomal location of eleven Elytrigia elongata (= Agropyron elongatum) isozyme structural genes. Genet Res 41:181–202
Karpechenko GD (1922) The number of chromosomes and the genetic correlation of cultivated Cruciferae. Bull Appl Bot Genet Plant Breed 13:3–14
Keller WA, Armstrong KC (1983) Production of haploids via anther culture in Brassica oleracea var. italica. Euphytica 32:151–159
Khush GS (1973) Genetics of aneuploids. Academic Press, New York, 301 pp
Landry BS, Michelmore RW (1985) Selection of probes for restriction fragment length analysis from plant genomic clones. Plant Mol Biol Rep 3:174–179
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning, a laboratory manual. Cold Springs Harbor Laboratory, Cold Springs Harbor, New York, 545 pp
Morinaga T (1934) On the chromosome number of Brassica juncea and B. napus, on the hybrid between the two, and on offspring line of the hybrid. Jpn J Genet 9:161–163
Mizushima U (1980) Genome analysis in Brassica and allied genera. In: Tsunoda S, Hinata K, Gomez-Campos C (eds) Brassica crops and wild allies. Japan Scientific Societies Press, Tokyo, pp 89–106
Nitsch JP, Nitsch C (1969) Haploid plants from pollen grains. Science 163:85
Palmer JD, Shields CR, Cohen DB, Orton TJ (1983) Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor Appl Genet 65:181–189
Peffley EB, Corgan JN, Horak KE, Tanksley SD (1985) Electrophoretic analysis of Allium alien addition lines. Theor Appl Genet 71:176–184
Prakash S, Hinata K (1980) Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot 55:1–57
Quiros CF, McHale N (1985) Genetic analysis of isozyme variants in diploid and tetraploid potatoes. Genetics 111:131–145
Quiros CF, Kianian SF, Ochoa O, Douches D (1985) Genome evolution in Brassica: use of molecular markers and cytogenetic stocks. Cruciferae Newslett 10:21–23
Riley R, Chapman V, Kimber G (1959) Genetic control of chromosome pairing in intergeneric hybrids with wheat. Nature 183:1244–1245
Robbelen G (1960) Beiträge zur Analyse des Brassica-Genomes. Chromosoma 11:205–228
Robbins MP, Vaughan JG (1983) Rubisco in Brassicaceae. In: Jensen U, Fairbrothers DE (eds) Proteins and nucleic acids in plant systematics. Springer, Berlin, pp 191–204
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. PNAS USA 81:8014–8018
Sears ER (1969) Wheat cytogenetics. Annu Rev Genet 3:451–468
Sikka SM (1940) Cytogenetics of Brassica hybrids and species. J Genet 40:441–509
Stebbins GL (1971) Chromosomal evolution in higher plants. Addison-Wesley, California
Swaminathan MS, Magoon ML, Mehra KL (1954) A simple propionic-carmine PMC smear method for plants with small chromosomes. Ind J Genet Plant Breed 14:87–88
U, N (1935) Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Genet 7:389–452
Weeden NF, Gottlieb LD (1980) Isolation of cytoplasmic enzymes from pollen. Plant Physiol 66:400–403
Williams PH, Hill CB (1986) Rapid-cycling populations of Brassica. Science 232:1385–1389
Author information
Authors and Affiliations
Additional information
Communicated by C.S. Khush
Rights and permissions
About this article
Cite this article
Quiros, C.F., Ochoa, O., Kianian, S.F. et al. Analysis of the Brassica oleracea genome by the generation of B. campestris-oleracea chromosome addition lines: characterization by isozymes and rDNA genes. Theoret. Appl. Genetics 74, 758–766 (1987). https://doi.org/10.1007/BF00247554
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00247554