Abstract
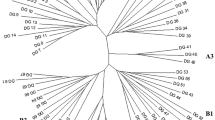
It has frequently been suggested to use the resynthesis of rapeseed (Brassica napus) from B. campestris and B. oleracea to broaden its genetic base. The objective of the present study is twofold: (1) to compare the genetic variation within resynthesized rapeseed with a world-wide collection of oilseed rape cultivars, and (2) to compare genetic distances estimated from RFLP markers with distances estimated from a relatively small number of allozyme markers. We investigated 17 resynthesized lines and 24 rapeseed cultivars. Genetic distances were estimated either based on the electrophoresis of seven allozymes, with a total of 38 different bands, or based on RFLP data of 51 probe/enzyme combinations, with a total of 355 different bands. The results of allozyme and RFLP analyses agreed reasonably well. Genetic distances, estimated from two independent sets of RFLP data with 25 and 26 probe/enzyme combinations respectively, were highly correlated; hence about 50 RFLP markers are sufficient to characterize rapeseed material with a large genetic diversity. The cultivars were clustered into three groups: (1) spring rapeseed of European and Northern American origin, (2) winter rapeseed of European and Northern American origin, and (3) rapeseed of Asian origin. Several of the resynthesized rapeseed lines were similar to European winter rapeseed cultivars, whereas others had quite unique patterns. It is concluded, that resynthesized rapeseed is a valuable source for broadening the genetic variation in present breeding material of Brassica napus. However, different lines differ widely in their suitability for this purpose.
Similar content being viewed by others
References
Akbar MA (1989) Chromosomal stability and performance of resynthesized Brassica napus produced for gain in earliness and shortday response. Hereditas 111:247–253
Andersson G, Olsson G (1961) Cruciferen — Ölpflanzen. In: Kappert H, Rudorf W (eds) Handbuch der Pflanzenzüchtung, 2. Aufl., Band V. Parey, Berlin, pp 1–66
Becker HC, Damgaard C, Karlsson B (1992) Environmental variation for outcrossing rate in rapeseed (Brassica napus). Theor Appl Genet 84:303–306
Boppenmaier J, Melchinger AE, Seitz G, Geiger HH, Hermann RG (1993) Genetic diversity for RFLPs in European maize inbreds. III. Performance of crosses within versus between heterotic groups for grain traits. Plant Breed 111:217–226
Chen BY, Heneen WK (1989) Resynthesized Brassica napus L.: A review of its potential in breeding and genetic analysis. Hereditas 111:255–263
Chen BY, Heneen WK, Jönsson R (1988) Resynthesis of Brassica napus L. through interspecific hybridization between B. alboglabra Bailey and B. campestris L. with special emphasis on seed colour. Plant Breed 101:52–59
Clegg MT (1989) Molecular diversity in plant populations. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources. Sinauer, Sunderland, Massachusetts, pp 98–115
Diedrichsen E, Sacristan MD (1991) Resynthesis of amphidiploid Brassica species and their clubroot disease reactions. Proc GCIRC 1991 Congr 1:274–279
Engqvist GM, Becker HC (1994) What can resynthesized Brassica napus offer to plant breeding? Sver Utsädesförenings Tidsk 104:87–92
Gland A, Röbbelen G, Thies W (1981) Variation of alkenyl glucosinolates in seeds of Brassica species. Z Pflanzenzüchtg 87:96–110
Herbert PDN, Beaton MJ (1989) Methodologies for allozyme analysis using cellulose acetate electrophoresis. Helena Laboratories, Beaumont, Texas, USA
Krähling K (1987) Utilitzation of genetic variability of resynthesized rapeseed. Plant Breed 99:209–217
Link W, Dixkens C, Singh M, Schwall M, Melchinger AE (1995) Genetic diversity in European and Mediterranean faba bean germplasm revealed by RAPD markers. Theor Appl Genet 90:27–32
Liu Z, Furnier GR (1993) Comparison of allozyme, RFLP, and RAPD markers for revealing genetic variation within and between trembling aspen and bigtooth aspen. Theor Appl Genet 87:97–105
Mailer RJ, Scarth R, Fristensky B (1994) Discrimination among cultivars of rapeseed (Brassica napus L.) using DNA polymorphisms amplified from arbitrary primers. Theor Appl Genet 87:697–704
McGrawth JM, Quiros CF (1992) Genetic diversity at isozyme and RFLP loci in Brassica campestris as related to crop type and geographical origin. Theor Appl Genet 83:783–790
Messmer MM, Melchinger AE, Lee M, Woodman WL, Lee EA, Lamkey KR (1991) Genetic diversity among progenitors and elite lines from the Iowa Stiff Stalk Synthetic (BSSS) maize population: comparison of allozyme and RFLP data. Theor Appl Genet 83:97–107
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Olsson G (1960) Species crosses within the genus Brassica II. Artificial Brassica napus L. Hereditas 46:351–396
Olsson G, Ellerström S (1980) Polyploidy breeding in Europe. In: Tsunda S, Hinata K, Goméz-Campo C (eds) Brassica crops and wild allies. Biology and breeding. Jpn Sci Soc Press. Tokyo, pp 167–190
Rohlf FJ (1993) NTSYS-pc, version 1.80. Exeter Software, Setauket, New York, USA
Song KM, Osborn TC (1992) Polyphyletic origins of Brassica napus: new evidence based on organelle and nuclear RFLP analyses. Genome 35:992–1001
Song K, Osborn TC, Williams PH (1990) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). 3. Genome relationships in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. campestris). Theor Appl Genet 79:497–506
Song K, Tang K, Osborn TC (1993) Development of synthetic Brassica amphidiploids by reciprocal hybridization and comparison to natural amphidiploids. Theor Appl Genet 86:811–821
Zhang Q, Saghai-Maroof MA, Kleinhofs A (1993) Comparative diversity analysis of RFLPs and isozymes within and among populations of Hordeum vulgare ssp. spontaneum. Genetics 134:909–916
Author information
Authors and Affiliations
Additional information
Communicated by G. Wenzel
Rights and permissions
About this article
Cite this article
Becker, H.C., Engqvist, G.M. & Karlsson, B. Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theoret. Appl. Genetics 91, 62–67 (1995). https://doi.org/10.1007/BF00220859
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00220859