Abstract
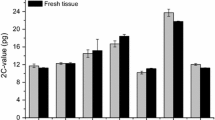
The nuclear DNA content of 23 selachian species (10 Batoidea, 11 Galeomorphii, and 2 Squalomorphii) was histophotometrically studied. Their genome sizes range from 7.5 pg/N in Raja fillae (Batoidea) to 34.1 pg/N in Oxynotus centrina (Squalomorphii).
Results show slight differences in the pattern of quantitative variations between the superorders Batoidea and Galeomorphii; Squalomorphii preserve their peculiar wide interspecific variability at the intrafamilial level, with values sited between 13.1 and 34.1 pg/N.
In 21 species also the DNA base composition was determined by means of DAPI. The study shows that in the species examined the DAPI positive fraction varies from a minimum of 27.7% in Oxynotus centrina, which possesses the largest genome size among all the Selachians studied, to a maximum of 72.5% in Carcharhinus limbatus. As a whole the data show an inverse correlation between the DNA content and the DAPI positive fraction, a condition common to all cold-blooded vertebrates.
The low percentage of DAPI positive DNA found in Oxynotus centrina could be attributable to a lower stainability by the fluorochrome caused by a higher chromatin condensation in the erythrocytes.
The validity of the DAPI method was verified by comparison with the biochemical assay according to the thermal denaturation method in 6 selachian species.
Similar content being viewed by others
References
Capriglione, T., Olmo, E., Odierna, G., Improta, B. & Morescalchi, A., 1987. Cytofluorometric DNA base determination in vertebrate species with different genome sizes. Bas. appl. Histochem. 31: 119–126.
Coleman, A. W. & Goff, L. J., 1985. Applications of fluorochromes to pollen biology. I. Mythramycin and 4′,6′-diamidino-2-phenylindole (DAPI) as vital stains and for quantitation of nuclear DNA. Stain Technol. 60: 145–154.
Hinegardner, R. T., 1976. The cellular DNA content of sharks, rays and some other fishes. Comp. Biochem. Physiol. 55B: 367–370.
Leeman, U. & Ruch, F., 1982. Cytofluorometric determination of DNA base content in plant nuclei and chromosomes by the fluorochromes DAPI and chromomycin A3. Expl Cell Res. 140: 275–282.
Maisey, J. G., 1984a. Chondrichthyan phylogeny: a look at the evidence. J. Vert. Pal. 4: 359–371.
Maisey, J. G., 1984b. Higher elasmobranch phylogeny and biostratigraphy. Zool. J. Linn. Soc. 82: 33–54.
Mandel, M. & Marmur, J., 1968. Use of ultraviolet absorbance-temperature profile for determining the Guanine plus Cytosine content of DNA. Methods Enzymol. 12B, 195–206.
Marmur, J., 1963. A procedure for the isolation of deoxyribonucleic acid from microorganisms. Methods Enzymol. 6: 726–738.
Mendelsohn, M. L., 1966. Adsorption cytophotometry: comparative methodology for objects and the two-wavelength method. In: Introduction to quantitative cytochemistry: 201–214, G. L.Wied, ed., Academic Press, New York and London.
Nagl, W., 1985. Chromatin organization and the control of gene activity. Int. Rev. Cytol. 94: 21–56.
Olmo, E., 1983. Nucleotype and cell size in vertebrates: a review. Bas. appl. Histochem. 27: 227–256.
Olmo, E., Stingo, V., Odierna, G. & Capriglione, T., 1980. Cryptic polyploidy in sharks and rays as revealed by DNA renaturation Kinetics. Atti Accad. Naz. Line. 48: 555–560.
Olmo, E., Stingo, V., Cobror, O., Capriglione, T. & Odierna, G., 1982. Repetitive DNA and polyplody in selachians. Comp. Biochem. Physiol. 73b: 739–745.
Otto, F. & Tsou, K. C., 1985. A comparative study of DAPI, DIPI and Hoechst 33258 and 33342 as chromosomal DNA stains. Stain Technol. 60: 7–11.
Schaeffer, B., 1967. Comments on elasmobranch evolution. In: Sharks, skates and rays. P. W.Gilbert, R. F.Mathewson and D. P.Rall, eds. John Hopkins Press, Baltimore.
Schwartz, F. J. & Maddock, M. B., 1986. Comparisons of karyotypes and cellular DNA contents within and between major lines of elasmobranchs. In: Indo-Pacific fish Biology: 148–157, T.Uyeno, R.Arai, T.Taniuchi & K.Matsuura, eds, Ichthyol. Soc. Jap., Tokio.
Stingo, V., DuBuit, M. H. & Odierna, G., 1980. The genome size of some selachian fishes. Boll. Zool. 47: 129–137.
Stingo, V. & Capriglione, T., 1986. DNA and chromosomal evolution in cartilaginous fish. In: Indo-Pacific fish Biology: 140–147, T.Uyeno, R.Arai, T.Taniuchi & K.Matsuura, eds, Ichthyol. Soc. Jap., Tokio.
Stingo, V., 1986. Correlazioni tra quantità di DNA totale e DNA DAPI-positivo in Teleostei e Selaci. Bas. appl. Histochem., 30/suppl., 164.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Stingo, V., Capriglione, T., Rocco, L. et al. Genome size and A-T rich DNA in selachians. Genetica 79, 197–205 (1989). https://doi.org/10.1007/BF00121513
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00121513