Abstract
Navigation, acquisition and interpretation of ultrasound (US) images relies on the skills and expertise of the performing physician. Virtual-reality based simulations offer a safe, flexible and standardized environment to train these skills. Simulations can be data-based by displaying a-priori acquired US volumes, or ray-tracing based by simulating the complex US interactions of a geometric model. Here we combine these two approaches as it is relatively easy to gather US images of normal background anatomy and attractive to cover the range of rare findings or particular clinical tasks with known ground truth geometric models. For seamless adaption and change of US content we further require stitching, texture synthesis and tissue deformation simulations. We test the proposed hybrid simulation method by replacing embryos within gestational sacs by ray-traced embryos, and by simulating an ectoptic pregnancy.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Due to portability, low-costs and safety, ultrasound (US) is a widely-used medical imaging modality. A drawback of US is that its acquisition and interpretation heavily relies on the experience and skill of the sonographer. Therefore, training of sonographers in navigation, acquisition and interpretation of US images is crucial for the benefit of clinical outcomes. Training is possible with volunteers, cadavers, and phantoms, which all have associated ethical and realism issues. On the other hand, virtual-reality based simulated training offers a safe and repeatable environment. This also allows to simulate rare cases, which would otherwise be unlikely to be encountered during training in regular clinical routine [2].
Real-time US simulations can be performed using data-based approaches [1, 6, 9, 15, 18, 19] or model-based rendering approaches [4, 8, 13, 16, 17]. Data-based US simulation can provide relatively high image realism, where image slices are interpolated during simulation time from a-priori acquired US volumes, which can also incorporate interactive tissue deformations [9]. While the acquisition of a large image database for physiological cases may seem straightforward, their preprocessing, storage, and evaluation-metric definition for simulation can quickly become infeasible. Furthermore, acquisition of rare cases with representative diversity, which are indeed most important for training, is a major challenge. For instance, for a comprehensive training in obstetrics, it is infeasible to collect US volumes of fetuses in all gestational ages, at different position/orientations, with different (uterus) anatomical variations, and all combinations thereof with a standardized image quality. If one wants to include cases of various imaging failures and artifact combinations, let alone rare pathological scenarios such as Siamese twins, the challenges for comprehensive training becomes apparent.
Alternatively, model-based techniques allow generating US images from a user set model, such as ray-tracing techniques through triangulated surface models of anatomy. These are then not limited by acquisition, but rather the modeling effort; i.e., any anatomical variation that can be modeled can be simulated. Nevertheless, precise modeling is a time-consuming effort and can only be afforded at small regions of discernible detail and actual clinical interest. For instance, [13] shows that a fetus (which has relatively small anatomical differences across its population) can be modeled in detail based on anatomical literature and expertise, however, it is clearly infeasible to model large volumes of surrounding background (mother’s) anatomy, e.g. the intestines, and their population variability for diverse realistic US backgrounds for comprehensive training.
In this work, we propose to combine these two approaches, where a focal region of clinical interest can be modeled in detail, which is then fused with realistic image volumes forming the background. A framework (Fig. 1) and related tools are then herein introduced for synthesizing realistic US images for different simulation scenarios, where, for instance, the original images may contain content that needs to be removed (“erased”) or the models may be smaller/larger than the available space for them in the images. A toolbox of hybrid US synthesis is demonstrated in this work. The proposed methods are showcased for two representative transvaginal ultrasound (TVUS) training scenarios: (i) regular fetal examination (of normal pregnancies, e.g., for controlling normative development and gestational age), and (ii) diagnosing ectopic pregnancies, which occurs when a fertilized ovum implants outside the normal uterine cavity and may lead, untreated in severe cases, to death.
2 Methods
2.1 Data-Based: Realistic Background from Images
Routine clinical images can be used to provide realistic examples for most of the anatomy. Here a mechanically-swept 3D TVUS transducer was used to collect image volumes during obstetric examination of 3 patients. Anatomical relevant structures were then manually annotated for further processing of the images and for placement and alignment of the model with the US volume. For the shown normal pregnancy scenarios, embryo, yolk sac and gestational sac were segmented, and the location of the umbilical cord at the placenta was marked.
2.2 Model-Based Simulation: Detailed, Arbitrary Content
New content is simulated from surface models using a Monte-Carlo ray tracing method [12, 13]. In this method, scatterers are handled using a convolution-based US simulation, while large-scale structures (modeled surfaces) are handled using ray-tracing, ignoring the wave effects. Scatters were parameterization at a fine and coarse level using normal distributions (\(\mu _s, \sigma _s\)), and a density (\(\rho _s\)) which gives a probability threshold above which a scatterer will provide an image contribution (i.e. convolution will be performed). Monte-Carlo ray-tracing allowes to simulate tissue interactions, like soft shadows and fuzzy reflections. Furthermore it makes it possible to parameterize surface properties, such as interface roughness (random variation of reflected/refracted ray direction) and interface thickness. Probe and tissue configurations, as well as rendering parameters were set according to the work in [13]. Three hand-crafted embryo models with increasing anatomical complexity and crown-rump lengths, respectively, of 10, 28, and 42 mm were used to represent gestational ages 7, 9.5, and 11 weeks [11], with the largest model consisting of \(\approx \)1 million triangles, taking <1 s for ray-tracing.
2.3 Tissue Deformations
Incorporating new content can require to remove or create space, e.g. to simulate growth of the fetus. The surrounding tissue should accordingly deform. We simulated a homogeneous increase of the empty (zero intensity) gestational sac by dilating its mask with a spherical structure element of radius r. To get a deformation which has a controllable amount of strain, we used the signed distance map with respect to the new structure. We defined the motion magnitude \(m(\mathbf {x})\) from distance map \(D(\mathbf {x})\) by
where s is the scaling factor. The direction of the displacement field is given by the gradient of \(D(\mathbf {x})\), see Fig. 2. We scaled r by 1.1 to ensure that the dilated structure is filled with zero intensities and used \(s=0.1\).
2.4 Texture Filling
Regions consisting of homogeneous tissue, like the gestational sac, might be simulated by texture filling. We employ the method from Efros and Leung [5] for texture filling, as it performed significantly better than other methods for reproducing homogeneous tissue regions in B-mode US images [14]. It is based on filling an image region iteratively starting at its border by matching the intensity of valid voxels in border patches to exemplar patches.
Texture filling is organized via 3 mask images, namely \(M_f\), \(M_v\) and \(M_p\). For target image I, voxels to be filled are indicated by  and valid voxels which should not be changed are marked by \(M_v>0\). Exemplar patches of size \(7\times 7\times 7\) are extracted from regions where \(M_p>0\) for all patch voxels from source image P.
and valid voxels which should not be changed are marked by \(M_v>0\). Exemplar patches of size \(7\times 7\times 7\) are extracted from regions where \(M_p>0\) for all patch voxels from source image P.
For filling the gestational sac inclusive the embryo, P is the original image and patches are extracted from inside the gestational sac (\(M_g\)) excluding the embryo (\(M_e\)): \(M_p=M_g-M_e\). The target image I is also the original image, which should be filled inside the gestational sac including the embryo, i.e. \(M_f=M_g+M_e\), and which has valid voxels inside the US field of view (FOV) excluding \(M_g\) and \(M_e\).
Filling starts with non-filled voxels at the border of \(M_v\), i.e. \(B=\{\mathbf {x}\,|\,M_f(\mathbf {x})>0\), \(M_v(\mathbf {y})>0\}\), where \(\mathbf {y}\) is in the 26-connected 3D neighborhood of \(\mathbf {x}\). Patches centered at these border voxels B are processed in descending order of their numbers of valid voxels. For each border voxel \(\mathbf {x}\in B\), its surrounding patch is compared to all the example patches using sum of squared differences (SSD). Candidate patches for filling the current voxel are those with SSD of all valid voxel values below a given threshold \(\theta \). If no matching patch is found, \(\theta \) is increased by 10%. From these candidates only those with SSD smaller than the minimal \({SSD}_{min}\) plus a given tolerance (\(1.3\,{SSD}_{min}\)) are accepted. Then, one of the accepted patches is randomly chosen and the intensity of its central voxel is assigned to the border voxel \(\mathbf {x}\). Finally the masks \(M_v\) and \(M_f\) are updated by setting \(M_v(\mathbf {x})=1\) and \(M_f(\mathbf {x})=0\). The above is iterated until all voxels are filled. As our 3D implementation runs very slow and the mechanically-swept probe anyhow collects volumes slice-wise, we performed texture filling slice-wise in 2D for all slices which include \(M_f\).
2.5 Compounding Contents
Overlapping content of US volumes (real or simulated) was combined by stitching [7]. This preserves speckle patterns and avoids image blurring/degradation of common mean/median approaches by determining a cut interface, such that each voxel comes from a single volume. This interface is found by capturing the transition quality between neighboring voxels by edge potential in a graphical model, which is then optimized via graph-cut [10]. In details, neighboring voxels \(\mathbf {x}\) and \(\mathbf {y}\) in overlapping volumes \(V_1\) and \(V_2\) have edge potential p based on their image intensity and image gradients [7]:
where \(\nabla ^e_{V_i}\) is the gradient in \(V_i\) along the graph edge e and \(\epsilon =10^{-5}\) to avoid division by zero. This encourages cutting when intensities match (numerator small) and at image edges (denumerator large) where seams are likely not visible. A graph G is constructed for only the overlapping voxels, with source s or sink t of the graph being connected to all boundary voxels of the corresponding image. Finally, the minimum cost cut of this graph is found using [3], giving a partition of G such that \(\min \sum _{\mathbf {x}\in V_1,\mathbf {y}\in V_2|e=(\mathbf {x},\mathbf {y})\in E}p(\mathbf {x},\mathbf {y}).\)
Even with optimal cuts, there can still be artifacts along stitched interfaces where no suitable seams exists, e.g. due to a quite small overlap and view-dependent artifacts like shadows. We reduced these artifacts by blending the volumes across the seam using a sigmoid function with a small kernel \(\sigma =3\) voxels.
3 Results
There is a wide range of potential applications for the proposed US hybrid simulation framework. We demonstrate its usefulness on four examples.
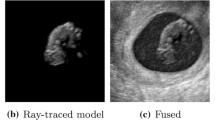
Case A: Normal Pregnancy, Similar size, Similar location. (Fig. 3) Replacement of a 10 week embryo by a 9 week model with know dimensions. Model placement required removal of real embryo, texture synthesis and stitching. Boundaries are clearly visible for simple fusion, which disappear with stitching.
Case B: Normal Pregnancy, Similar size, Different Location. (Fig. 4) Illustration of placing the 9 week embryo model at a different location for the same patient as in case A. High quality texture synthesis is required to realistically fill the regions where the real embryo was.
Case C: Normal Pregnancy, Simulation of Growth. (Fig. 5) A two week development of a 9 week embryo was simulated. This requires all components of the proposed framework including deformation simulation. Challenges include creation of a smooth deformation and realistic speckle patterns within and on the boundary of the gestational sac.
Case D: Ectopic Pregnancy. (Fig. 6) As abnormality we simulated an ectopic pregnancy. Guided by an US specialist in obstetrics and gynecology, we replaced normal tissue close to the ovaries by the model of a 7 week embryo and its gestational sac. Simulation parameters were set empirically for visually best matching of speckle pattern to the surrounding, with resulting image realism confirmed qualitatively by an sonographer in gynecology.
4 Conclusions
We propose a hybrid ultrasound simulation framework, where particular anatomy including rare cases is generated from anatomical models, while normal variability is covered by fusing it with real image data to reduce modeling efforts. Successful combination of these two data sources has been demonstrated for four cases within the context of obstetric examinations. Computations took \({<}\)10 mins. Volumes fused offline can be used in real-time image-based simulation, e.g. [9].
References
Arkhurst, W., et al.: A virtual reality training system for pediatric sonography. Int. Congress Ser. 1230, 483–487 (2001)
Blum, T., Rieger, A., Navab, N., Friess, H., Martignoni, M.: A review of computer-based simulators for ultrasound training. Simul. Healthcare 8, 98–108 (2013)
Boykov, Y., Kolmogorov, V.: An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans. Pattern Anal. Mach. Intell. 26(9), 1124–1137 (2004)
Bürger, B., Bettinghausen, S., Radle, M., Hesser, J.: Real-time GPU-based ultrasound simulation using deformable mesh models. IEEE Trans. Med. Imaging 32(3), 609–618 (2013)
Efros, A., Leung, T.: Texture synthesis by non-parametric sampling. In: IEEE International Conference on Computer Vision (ICCV), pp. 1033–1038 (1999)
Ehricke, H.: SONOSim3D: a multimedia system for sonography simulation and education with an extensible case database. Eur. J. Ultrasound 7(3), 225–300 (1998)
Flach, B., Makhinya, M., Goksel, O.: PURE: panoramic ultrasound reconstruction by seamless stitching of volumes. In: Tsaftaris, S.A., Gooya, A., Frangi, A.F., Prince, J.L. (eds.) SASHIMI 2016. LNCS, vol. 9968, pp. 75–84. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46630-9_8
Gao, H., et al.: A fast convolution-based methodology to simulate 2-D/3-D cardiac ultrasound images. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 56(2), 404–409 (2009)
Goksel, O., Salcudean, S.E.: B-mode ultrasound image simulation in deformable 3-D medium. IEEE Trans. Med. Imaging 28(11), 1657–1669 (2009)
Kwatra, V., Schödl, A., Essa, I., Turk, G., Bobick, A.: Graphcut textures: image and video synthesis using graph cuts. ACM Trans. Graph. (ToG). 22, 277–286 (2003)
Loughna, P., Chitty, L., Evans, T., Chudleigh, T.: Fetal size and dating: charts recommended for clinical obstetric practice. Ultrasound 17(3), 160–166 (2009)
Mattausch, O., Goksel, O.: Monte-Carlo ray-tracing for realistic interactive ultrasound simulation. In: Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM), pp. 1–9, Bergen, Norway, September 2016. https://doi.org/10.2312/vcbm.20161285
Mattausch, O., Makhinya, M., Goksel, O.: Realistic ultrasound simulation of complex surface models using interactive Monte-Carlo path tracing, vol. 37, pp. 202–213 (2018)
Mattausch, O., Ren, E., Bajka, M., Vanhoey, K., Goksel, O.: Comparison of texture synthesis methods for content generation in ultrasound simulation for training. In: SPIE Medical Imaging, p. 1013523 (2017)
Maul, H., et al.: Ultrasound simulators: experience with the SonoTrainer and comparative review of other training systems. Ultrasound Obstet. Gynecol. 24(5), 581–585 (2004)
Reichl, T., Passenger, J., Acosta, O., Salvado, O.: Ultrasound goes GPU: real-time simulation using CUDA. In: SPIE Medical Imaging, p. 726116 (2009)
Salehi, M., Ahmadi, S.-A., Prevost, R., Navab, N., Wein, W.: Patient-specific 3D ultrasound simulation based on convolutional ray-tracing and appearance optimization. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9350, pp. 510–518. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24571-3_61
Sclaverano, S., Chevreau, G., Vadcard, L., Mozer, P., Troccaz, J.: BiopSym: a simulator for enhanced learning of ultrasound-guided prostate biopsy. Stud. Health Technol. Inform. 142, 301–306 (2009)
Tahmasebi, A.M., Abolmaesumi, P., Hashtrudi-Zaad, K.: A haptic-based ultrasound training/examination system (HUTES). In: IEEE International Conference on Robotics and Automation (ICRA), pp. 3130–3131 (2007)
Acknowledgment
Funding was provided by Innosuisse Swiss Innovation Agency.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Tanner, C., Starkov, R., Bajka, M., Goksel, O. (2018). Framework for Fusion of Data- and Model-Based Approaches for Ultrasound Simulation. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11073. Springer, Cham. https://doi.org/10.1007/978-3-030-00937-3_39
Download citation
DOI: https://doi.org/10.1007/978-3-030-00937-3_39
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00936-6
Online ISBN: 978-3-030-00937-3
eBook Packages: Computer ScienceComputer Science (R0)