Abstract
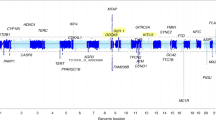
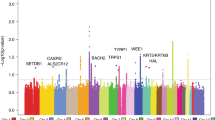
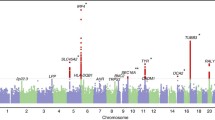
Chromosome 9p21 has been implicated in the pathogenesis of cutaneous malignant melanoma (CMM). In addition to CDKN2A, the major known high-risk susceptibility gene for CMM, recent studies suggest that other 9p21 genes may be involved in melanoma/nevi development. To identify 9p21 variants that influence susceptibility to CMM and number of nevi in CMM-prone families with and without CDKN2A mutations, we analyzed 562 individuals (183 CMM) from 53 families (23 CDKN2A+, 30 CDKN2A−) for 233 tagging SNPs in 21 genes at 9p21. Single SNP- and gene-based regression analyses were used to assess the risk of CMM, nevi count, skin complexion, and tanning ability associated with these SNPs and genes. We found that SNP rs7023329 in the MTAP gene was associated with number of nevi (P trend = 0.003) confirming a recent finding by a genome-wide association study. In addition, three SNPs in the ACO1 gene, rs7855483 (P trend = 0.002), rs17288067 (P trend = 0.0009), and rs10813813 (P trend = 0.005), showed the strongest associations with CMM risk. None of the examined 9p21 SNPs was associated with skin complexion, whereas two SNPs, rs10964862 in IFNW1 (P trend = 0.003), and rs13290968 in TUSC1 (P trend = 0.0006), were associated with tanning ability. Gene-based analyses suggested that the ACO1 gene was significantly associated with CMM (P = 0.0004); genes IFNW1 (P = 0.002) and ACO1 (P = 0.0002) were significantly associated with tanning ability. Our findings are consistent with recent proposals that additional 9p21 genes may contribute to CMM susceptibility in CMM-prone families. These genetic variants may, at least partially, exert their effects through nevi and tanning ability.

Similar content being viewed by others
Abbreviations
- CMM:
-
Cutaneous malignant melanoma
- SNP:
-
Single nucleotide polymorphism
- GWAS:
-
Genome-wide association study
- MAF:
-
Minimum minor allele frequency
- OR:
-
Odds ratio
- 95% CI:
-
95% Confidence interval
- GEE:
-
Generalized estimating equations
References
Tucker MA, Goldstein AM (2003) Melanoma etiology: where are we? Oncogene 22(20):3042–3052
Eliason MJ, Larson AA, Florell SR et al (2006) Population-based prevalence of CDKN2A mutations in Utah melanoma families. J Invest Dermatol 126(3):660–666
Goldstein AM (2004) Familial melanoma, pancreatic cancer and germline CDKN2A mutations. Hum Mutat 23(6):630
Rakosy Z, Vizkeleti L, Ecsedi S et al (2008) Characterization of 9p21 copy number alterations in human melanoma by fluorescence in situ hybridization. Cancer Genet Cytogenet 182(2):116–121
Zhu G, Montgomery GW, James MR et al (2007) A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur J Hum Genet 15(1):94–102
Falchi M, Spector TD, Perks U, Kato BS, Bataille V (2006) Genome-wide search for nevus density shows linkage to two melanoma loci on chromosome 9 and identifies a new QTL on 5q31 in an adult twin cohort. Hum Mol Genet 15(20):2975–2979
Liu L, Goldstein AM, Tucker MA et al (1997) Affected members of melanoma-prone families with linkage to 9p21 but lacking mutations in CDKN2A do not harbor mutations in the coding regions of either CDKN2B or p19ARF. Genes Chromosomes Cancer 19(1):52–54
Casula M, Ascierto PA, Cossu A et al (2003) Mutation analysis of candidate genes in melanoma-prone families: evidence of different pathogenetic mechanisms at chromosome 9P21. Melanoma Res 13(6):571–579
FitzGerald MG, Harkin DP, Silva-Arrieta S et al (1996) Prevalence of germ-line mutations in p16, p19ARF, and CDK4 in familial melanoma: analysis of a clinic-based population. Proc Natl Acad Sci USA 93(16):8541–8545
Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I (2007) Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67(8):3963–3969
Bishop DT, Demenais F, Iles MM et al (2009) Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet 41(8):920–925
Falchi M, Bataille V, Hayward NK et al (2009) Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet 41(8):915–919
Stevens AP, Spangler B, Wallner S et al (2009) Direct and tumor microenvironment mediated influences of 5′-deoxy-5′-(methylthio)adenosine on tumor progression of malignant melanoma. J Cell Biochem 106(2):210–219
Kirkwood JM, Strawderman MH, Ernstoff MS, Smith TJ, Borden EC, Blum RH (1996) Interferon alfa-2b adjuvant therapy of high-risk resected cutaneous melanoma: the Eastern Cooperative Oncology Group Trial EST 1684. J Clin Oncol 14(1):7–17
Goldstein AM, Landi MT, Tsang S, Fraser MC, Munroe DJ, Tucker MA (2005) Association of MC1R variants and risk of melanoma in melanoma-prone families with CDKN2A mutations. Cancer Epidemiol Biomark Prev 14(9):2208–2212
Goldstein AM, Struewing JP, Chidambaram A, Fraser MC, Tucker MA (2000) Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst 92(12):1006–1010
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74(1):106–120
Pfeiffer RM, Gail MH, Pee D (2001) Inference for covariates that accounts for ascertainment and random genetic effects in family studies. Biometrika 88:16
Zeger SL, Liang KY (1986) Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42(1):121–130
Allen-Brady K, Wong J, Camp NJ (2006) PedGenie: an analysis approach for genetic association testing in extended pedigrees and genealogies of arbitrary size. BMC Bioinformatics 7:209
Dudbridge F, Koeleman BP (2003) Rank truncated product of P-values, with application to genomewide association scans. Genet Epidemiol 25(4):360–366
Gandini S, Sera F, Cattaruzza MS et al (2005) Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer 41(1):28–44
Olopade OI, Pomykala HM, Hagos F et al (1995) Construction of a 2.8-megabase yeast artificial chromosome contig and cloning of the human methylthioadenosine phosphorylase gene from the tumor suppressor region on 9p21. Proc Natl Acad Sci USA 92(14):6489–6493
Hori Y, Hori H, Yamada Y et al (1998) The methylthioadenosine phosphorylase gene is frequently co-deleted with the p16INK4a gene in acute type adult T-cell leukemia. Int J Cancer 75(1):51–56
Wong YF, Chung TK, Cheung TH, Nobori T, Chang AM (1998) MTAP gene deletion in endometrial cancer. Gynecol Obstet Invest 45(4):272–276
Christopher SA, Diegelman P, Porter CW, Kruger WD (2002) Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res 62(22):6639–6644
Garcia-Castellano JM, Villanueva A, Healey JH et al (2002) Methylthioadenosine phosphorylase gene deletions are common in osteosarcoma. Clin Cancer Res 8(3):782–787
Behrmann I, Wallner S, Komyod W et al (2003) Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am J Pathol 163(2):683–690
Kadariya Y, Yin B, Tang B et al (2009) Mice heterozygous for germ-line mutations in methylthioadenosine phosphorylase (MTAP) die prematurely of T-cell lymphoma. Cancer Res 69(14):5961–5969
Toyokuni S (1996) Iron-induced carcinogenesis: the role of redox regulation. Free Radic Biol Med 20(4):553–566
Huang X (2003) Iron overload and its association with cancer risk in humans: evidence for iron as a carcinogenic metal. Mutat Res 533(1–2):153–171
Pra D, Rech Franke SI, Pegas Henriques JA, Fenech M (2009) A possible link between iron deficiency and gastrointestinal carcinogenesis. Nutr Cancer 61(4):415–426
Stevens RG, Beasley RP, Blumberg BS (1986) Iron-binding proteins and risk of cancer in Taiwan. J Natl Cancer Inst 76(4):605–610
Selby JV, Friedman GD (1988) Epidemiologic evidence of an association between body iron stores and risk of cancer. Int J Cancer 41(5):677–682
Stevens RG, Graubard BI, Micozzi MS, Neriishi K, Blumberg BS (1994) Moderate elevation of body iron level and increased risk of cancer occurrence and death. Int J Cancer 56(3):364–369
Knekt P, Reunanen A, Takkunen H, Aromaa A, Heliovaara M, Hakulinen T (1994) Body iron stores and risk of cancer. Int J Cancer 56(3):379–382
Herrinton LJ, Friedman GD, Baer D, Selby JV (1995) Transferrin saturation and risk of cancer. Am J Epidemiol 142(7):692–698
Pantopoulos K (2004) Iron metabolism and the IRE/IRP regulatory system: an update. Ann NY Acad Sci 1012:1–13
Zhang KH, Tian HY, Gao X et al (2009) Ferritin heavy chain-mediated iron homeostasis and subsequent increased reactive oxygen species production are essential for epithelial-mesenchymal transition. Cancer Res 69(13):5340–5348
Boult J, Roberts K, Brookes MJ et al (2008) Overexpression of cellular iron import proteins is associated with malignant progression of esophageal adenocarcinoma. Clin Cancer Res 14(2):379–387
Holmstrom P, Gafvels M, Eriksson LC et al (2006) Expression of iron regulatory genes in a rat model of hepatocellular carcinoma. Liver Int 26(8):976–985
Baldi A, Lombardi D, Russo P et al (2005) Ferritin contributes to melanoma progression by modulating cell growth and sensitivity to oxidative stress. Clin Cancer Res 11(9):3175–3183
Tan MG, Kumarasinghe MP, Wang SM, Ooi LL, Aw SE, Hui KM (2009) Modulation of iron-regulatory genes in human hepatocellular carcinoma and its physiological consequences. Exp Biol Med (Maywood) 234(6):693–702
Chen G, Fillebeen C, Wang J, Pantopoulos K (2007) Overexpression of iron regulatory protein 1 suppresses growth of tumor xenografts. Carcinogenesis 28(4):785–791
Gu F, Qureshi AA, Niu T et al (2008) Interleukin and interleukin receptor gene polymorphisms and susceptibility to melanoma. Melanoma Res 18(5):330–335
Howell WM, Turner SJ, Bateman AC, Theaker JM (2001) IL-10 promoter polymorphisms influence tumour development in cutaneous malignant melanoma. Genes Immun 2(1):25–31
Nikolova PN, Pawelec GP, Mihailova SM et al (2007) Association of cytokine gene polymorphisms with malignant melanoma in Caucasian population. Cancer Immunol Immunother 56(3):371–379
Hanneman KK, Cooper KD, Baron ED (2006) Ultraviolet immunosuppression: mechanisms and consequences. Dermatol Clin 24(1):19–25
Shan Z, Parker T, Wiest JS (2004) Identifying novel homozygous deletions by microsatellite analysis and characterization of tumor suppressor candidate 1 gene, TUSC1, on chromosome 9p in human lung cancer. Oncogene 23(39):6612–6620
Acknowledgments
We are indebted to the participating families, whose generosity and cooperation have made this study possible. We also acknowledge the contributions to this work that were made by Virginia Pichler, Deborah Zametkin, and Mary Fraser. This research was supported by the Intramural Research Program of the NIH, NCI, DCEG.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, X.R., Liang, X., Pfeiffer, R.M. et al. Associations of 9p21 variants with cutaneous malignant melanoma, nevi, and pigmentation phenotypes in melanoma-prone families with and without CDKN2A mutations. Familial Cancer 9, 625–633 (2010). https://doi.org/10.1007/s10689-010-9356-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-010-9356-3