Abstract
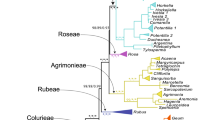
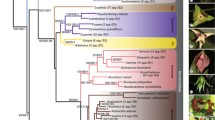
Phylogenetic relationships among 88 genera of Rosaceae were investigated using nucleotide sequence data from six nuclear (18S, gbssi1, gbssi2, ITS, pgip, and ppo) and four chloroplast (matK, ndhF, rbcL, and trnL-trnF) regions, separately and in various combinations, with parsimony and likelihood-based Bayesian approaches. The results were used to examine evolution of non-molecular characters and to develop a new phylogenetically based infrafamilial classification. As in previous molecular phylogenetic analyses of the family, we found strong support for monophyly of groups corresponding closely to many previously recognized tribes and subfamilies, but no previous classification was entirely supported, and relationships among the strongly supported clades were weakly resolved and/or conflicted between some data sets. We recognize three subfamilies in Rosaceae: Rosoideae, including Filipendula, Rubus, Rosa, and three tribes; Dryadoideae, comprising the four actinorhizal genera; and Spiraeoideae, comprising Lyonothamnus and seven tribes. All genera previously assigned to Amygdaloideae and Maloideae are included in Spiraeoideae. Three supertribes, one in Rosoideae and two in Spiraeoideae, are recognized.
Similar content being viewed by others
References
(2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 141: 399–436
Baillon H. (1869). Histoire des plantes, vol. 1. Librairie de L. Hachette, Paris
Benson D. R. and Silvester W. B. (1993). Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol. Rev. 57: 293–319
Bortiri E., Oh S., Gao F. and Potter D. (2002). The phylogenetic utility of nucleotide sequences of sorbitol 6-phosphate dehydrogenase in Prunus (Rosaceae). Amer. J. Bot. 89: 1697–1708
Bortiri E., Oh S., Jiang J., Baggett S., Granger A., Weeks C., Buckingham M., Potter D. and Parfitt D. (2001). Phylogeny and systematics of Prunus (Rosaceae) as determined by sequence analysis of ITS and the chloroplast trnL-trnF spacer DNA. Syst. Bot. 26: 797–807
Boss P. K., Gardner R. C., Janssen B. J. and Ross S. P. (1995). An apple polyphenol oxidase cDNA is up-regulated in wounded tissues. Pl. Molec. Biol. 27: 429–433
Campbell C. S., Donoghue M. J., Baldwin B. G. and Wojciechowski M. F. (1995). Phylogenetic relationships in Maloideae (Rosaceae): evidence from sequences of the internal transcribed spacers of nuclear ribosomal DNA and its congruence with morphology. Amer. J. Bot. 27: 903–918
Campbell C. S., Evans R. C., Morgan D. R., Dickinson T. A. and Arsenault M. P. (2007). Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Pl. Syst. Evol. 266: 119–145
Chevalier T., de Rigal D., Mbeguie-AMbeguie D., Gauillard F., Richard-Forget F., and Fils-Lycaon B. R. (1999). Molecular cloning and characterization of apricot fruit polyphenol oxidase. Pl. Physiol. (Lancaster) 119: 1261–1270
Chevreau E. and Laurens F. (1987). The pattern of inheritance in apple (Malus × domestica Borkh.): further results from leaf isozyme analysis. Theor. Appl. Genet. 75: 90–95
Chevreau E., Lespinasse Y. and Gallet M. (1985). Inheritance of pollen enzymes and polyploid origin of apple (Malus × domestica Borkh.). Theor. Appl. Genet. 71: 268–277
Cronquist A. (1981). An integrated system of classification of flowering plants. Columbia University Press, New York
Cuatrecasas J. (1970). Flora Neotropica Monograph No. 2, Brunelliaceae. Hafner, Darien, Connecticut
Du Mortier B.-C. (1827). Florula belgica: operis majoris prodromus. J. Casterman, Tournay
Eriksson T., Donoghue M. J. and Hibbs M. S. (1998). Phylogenetic analysis of Potentilla using DNA sequences of nuclear ribosomal internal transcribed spacers (ITS), and implications for the classification of Rosoideae (Rosaceae). Pl. Syst. Evol. 211: 155–179
Eriksson T., Hibbs M. S., Yoder A. D., Delwiche C. F. and Donoghue M. J. (2003). The phylogeny of Rosoideae (Rosaceae) based on sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA and the trnL/F region of chloroplast DNA. Int. J. Pl. Sci. 164: 197–211
Evans R. C. (1999) Molecular, morphological, and ontogenetic evaluation of relationships and evolution in the Rosaceae. Ph.D. dissertation, University of Toronto, Toronto.
Evans R. C. and Campbell C. S. (2002). The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. Amer. J. Bot. 89: 1478–1484
Evans R. C. and Dickinson T. A. (1999a). Floral ontogeny and morphology in subfamily Amygdaloideae T. and G. (Rosaceae). Int. J. Pl. Sci. 160: 955–979
Evans R. C. and Dickinson T. A. (1999b). Floral ontogeny and morphology in subfamily Spiraeoideae Endl. (Rosaceae). Int. J. Pl. Sci. 160: 981–1012
Evans R. C. and Dickinson T. A. (2005). Floral ontogeny and morphology in Gillenia (``Spiraeoideae'') and subfamily Maloideae C. Weber (Rosaceae). Int. J. Pl. Sci. 166: 427–447
Evans R. C., Alice L. A., Campbell C. S., Kellogg E. A. and Dickinson T. A. (2000). The granule-bound starch synthase (GBSSI) gene in the Rosaceae: multiple loci and phylogenetic utility. Molec. Phylogenet. Evol. 17: 388–400
Farr D. F. (1989). Fungi on plants and plant products in the United States. APS Press, St. Paul, Minnesota
Farr D.F.,Rossman A.Y., Palm M. E.,McCray E. B. (2005) Fungal databases, systematic botany and mycology laboratory. Agricultural Research Service, United States Department of Agriculture, available at http://nt.ars-grin.gov/fungaldatabases/.
Gladkova V. N. (1972). On the origin of subfamily Maloideae. Bot. Zhurn. 57: 42–49
Gray A. (1842). The botanical text-book. Putnam, New York
Greuter W., McNeil J., Barrie F. R., Burdet H. M., Demoulin V., Filgueiras T. S., Nicolson D. H., Silva P. C., Skog J. E., Trehane P., Turland N. J., Hawksworth D. L. (eds.) (2000) International code of botanical nomeclature. (Tokyo Code). Koeltz Scientific Books, Königstein.
Haruta M., Murata M., Hiraide A., Kadokura H., Yamasaki M., Sakuta M., Shimizu S. and Homma S. (1998). Cloning genomic DNA encoding apple polyphenol oxidase and comparison of the gene product in Escherichia coli and in apple. Biosci. Biotechnol. Biochem. 62: 358–362
Haruta M., Murata M., Kadokura H. and Homma S. (1999). Immunological and molecular comparison of polyphenol oxidase in Rosaceae fruit trees. Phytochemistry 50: 1021–1025
Helfgott D. M., Francisco-Ortega J, Santos-Guerra A., Jansen R. K. and Simpson B. B. (2000). Biogeography and breeding system evolution of the woody Bencomia alliance (Rosaceae) in Macaronesia based on ITS sequence data. Syst. Bot. 25: 82–97
Henrickson J. (1986). Notes on Rosaceae. Phytologia 468: 60
Huelsenbeck J. P. and Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755
Hutchinson J. (1964). The genera of flowering plants, vol. 1, Dicotyledons. Clarendon Press, Oxford
Hutchinson J. (1969). Evolution and phylogeny of flowering plants. Academic Press, London
International Plant Names Index (2006) Published on the internet at http://www.ipni.org.
Judd W. S. and Olmstead R. G. (2004). A survey of tricolpate (eudicot) phylogenetic relationships. Amer. J. Bot. 91: 1627–1644
Kalkman C. (1965). The Old World species of Prunus subgen. Laurocerasus including those formerly referred to Pygeum. Blumea 13: 1–115
Kalkman C. (2004). Rosaceae. In: Kubitzki, K. (eds) The families and genera of vascular plants, vol. 6, Flowering plants - Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales, pp 343–386. Springer, Berlin
Kerr M. S. (2004) A phylogenetic and biogeographic analysis of Sanguisorbeae (Rosaceae), with emphasis on the Pleistocene radiation of the high Andean genus Polylepis. Ph.D. dissertation, University of Maryland, College Park.
Koehne E. (1890). Die Gattungen der Pomaceen. Gaertner, Berlin
Kubitzki K. (2004). The families and genera of vascular plants, vol. 6, Flowering plants – Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales. Springer, Berlin
Lawrence G. H. M. (1951). Taxonomy of vascular plants. Macmillan, New York
Lee S. and Wen J. (2001). A phylogenetic analysis of Prunus and the Amygdaloideae (Rosaceae) using ITS sequences of nuclear ribosomal DNA. Amer. J. Bot. 88: 150–160
Mabberley D. J. (2002). Potentilla and Fragaria (Rosaceae) reunited. Telopea 9: 793–801
Maddison W. P. and Maddison D. R. (2003). MacClade, version 4.06. Analysis of phylogeny and character evolution. Sinauer Associates, Inc., Sunderland, Massachusetts
Missouri Botanical Garden (2005) Index to Plant Chromosome Numbers Database, available at http://mobot.mobot.org/W3T/Search/ipcn.html.
Morgan D. R., Soltis D. E. and Robertson K. R. (1994). Systematic and evolutionary implications of rbcL sequence variation in Rosaceae. Amer. J. Bot. 81: 890–903
Nylander J. A. A. (2005) MrAIC, version 1.4., available at http://www.abc.se/~nylander/.
Oh S. (2006). Neillia includes Stephanandra (Rosaceae). Novon 16: 91–95
Oh S. and Potter D. (2005). Molecular phylogenetic systematics and biogeography of tribe Neillieae (Rosaceae) using DNA sequences of cpDNA, rDNA and LEAFY. Amer. J. Bot. 92: 179–192
Oh S. and Potter D. (2006). Description and phylogenetic position of a new angiosperm family, Guamatelaceae, inferred from chloroplast rbcL, atpB and matK sequences. Syst. Bot. 31: 730–738
Pankhurst R. (2005) Rosaceae database. On-line searchable version available through the International Organization for Plant Information’s Provisional Global Plant Checklist at: http://bgbm3.bgbm.fu-berlin.de/iopi/gpc/query.asp.
Potter D. (2003). Molecular phylogenetic studies in Rosaceae. In: Sharma, A. K. and Sharma, A. (eds) Plant genome: Biodiversity and evolution, vol. I, Pt. A: Phanerogams, pp 319–351. Science Publishers, Inc. Enfield, New Hampshire
Potter D., Gao F., Bortiri P. E., Oh S. and Baggett S. (2002). Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnL-trnF nucleotide sequence data. Pl. Syst. Evol. 231: 77–89
Potter D., Still S. M., Ballian D. and Kraigher H. (2006). Phylogenetic relationships in tribe Spiraeeae (Rosaceae) inferred from nucleotide sequence data. Pl. Syst. Evol. 266: 105–118
Rambaut A. (1996) Se-Al: Sequence Alignment Editor. Available at http://evolve.zoo.ox.ac.uk/software.html.
Raspé O., Jacquemart A.-L. and De Sloover J. (1998). Isozymes in Sorbus aucuparia (Rosaceae: Maloideae): genetic analysis and evolutionary significance of zymograms. Int. J. Pl. Sci. 159: 627–636
Reveal J. L. (2004) Index nominum supragenericorum plantarum vascularum.?http://www. life.umd.edu/emeritus/reveal/pbio/WWW/supra gen.html.
Robertson K. R., Phipps J. B. and Rohrer J. R. (1991). A synopsis of genera in Maloideae (Rosaceae). Syst. Bot. 16: 376–394
Rohrer J. R., Robertson K. R. and Phipps J. B. (1994). Floral morphology of Maloideae (Rosaceae) and its systematic relevance. Amer. J. Bot. 81: 574–581
Roitman A., Flaks B. R., Fradkina L. Z. and Federov A. A. (1974). Chromosome numbers of flowering plants. Ger. Otto Koeltz Science Publishers, Koenigstein
Savile D. B. O. (1979). Fungi as aids in higher plant classification. Bot. Rev. 45: 380–495
Sax K. (1933). The origin of the Pomoideae. Proc. Amer. Soc. Hort. Sci. 30: 147–150
Schulze-Menz G. K. (1964). Rosaceae. In: Melchior, H. (eds) Engler's Syllabus der Pflanzenfamilien II, pp 209–218. Gebrüder Borntraeger, Berlin
Shaw J. and Small R. L. (2004). Addressing the “hardest puzzle in American pomology:” phylogeny of Prunus sect. Prunocerasus (Rosaceae) based on seven noncoding chloroplast DNA regions. Amer. J. Bot. 91: 985–996
Simpson C .G., Macrae E. and Gardner R. C. (1995). Cloning of a polygalacturonase inhibiting protein from kiwifruit (GenBank Z49063). Pl. Physiol. 108: 1748
Smedmark J. E. E. (2006). Recircumscription of Geum L. (Colurieae: Rosaceae). Bot. Jahrb. Syst. 126: 409–417
Smedmark J. E. E. and Eriksson T. (2002). Phylogenetic relationships of Geum (Rosaceae) and relatives inferred from the nrITS and trnL-trnF regions. Syst. Bot. 27: 303–317
Smedmark J. E. E., Eriksson T. and Bremer B. (2005). Allopolyploid evolution in Geinae (Colurieae: Rosaceae) – building reticulate species trees from bifurcating gene trees. Organisms Divers. Evolut. 5: 275–283
Smedmark J. E. E., Eriksson T., Evans R. C. and Campbell C. S. (2003). Ancient allopolypoloid speciation in Geinae (Rosaceae): evidence from nuclear granule-bound starch synthase (GBSSI) gene sequences. Syst. Biol. 52: 374–385
Soltis D. E., Soltis P. S., Chase M. W., Mort M. E., Albach D. C., Zanis M., Savolainen V., Hahn W. H., Hoot S. B., Fay M. F., Axtell M., Swensen S. M., Prince L. M., Kress W. J., Nixon K. C. and Farris J. S. (2000). Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 133: 381–461
Spjut R. W. (1994). A systematic treatment of fruit types. Mem. New York Bot. Gard. 70: 1–182
Staden R. (1996). The Staden sequence analysis package. Mol. Biotechnol. 5: 233–241
Sterling C. (1964). Comparative morphology of the carpel in the Rosaceae. III. Pomoideae: Crataegus, Hesperomeles, Mespilus, Osteomeles. Amer. J. Bot. 51: 705–712
Sterling C. (1965a). Comparative morphology of the carpel in the Rosaceae. IV. Pomoideae: Chamaemeles, Cotoneaster, Dichotomanthes, Pyracantha. Amer. J. Bot. 52: 47–54
Sterling C. (1965b). Comparative morphology of the carpel in the Rosaceae. V. Pomoideae: Amelanchier, Aronia, Malacomeles, Malus, Peraphyllum, Pyrus, Sorbus. Amer. J. Bot. 52: 418–426
Sterling C. (1965c). Comparative morphology of the carpel in the Rosaceae. VI. Pomoideae: Eriobotrya, Heteromeles, Photinia, Pourthiaea, Raphiolepis, Stranvaesia. Amer. J. Bot. 52: 938–946
Sterling C. (1966). Comparative morphology of the carpel in the Rosaceae. VII. Pomoideae: Chaenomeles, Cydonia, Docynia. Amer. J. Bot. 53: 225–231
Stotz H. U., Powell A. L. T., Damon S. E., Greve L. C., Bennett A. B. and Labavitch J. M. (1993). Molecular characterization of a polygalacturonase inhibitor from Pyrus communis L. cv. Bartlett. Pl. Physiol. (Lancaster) 102: 133–138
Stotz H. U., Contos J. J., Powell A. L., Bennett A. B. and Labavitch J. M. (1994). Structure and expression of an inhibitor of fungal polygalacturonases from tomato. Pl. Molec. Biol. 25: 607–617
Swofford D. L. (2002). PAUP* Phylogenetic analysis using (* and other methods) parsimony Version 4. Sinauer Associates, Sunderland, Massachusetts
Taberlet P., Gielly L., Patou G. and Bouvet J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl. Molec. Biol. 17: 1105–1109
Takhtajan A. (1997). Diversity and classification of flowering plants. Columbia University Press, New York
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. and Higgins D. G. (1997). The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 25: 4876–4882
Toubart P., Desiderio A., Salvi G., Cervone F., Daroda L. and De Lorenzo G. (1992). Cloning and characterization of the gene encoding the endopolygalacturonase-inhibiting protein (PGIP) of Phaseolus vulgaris L. Pl. J. 2: 367–373
Vanden Heuvel B. D., Benson D. R., Bortiri E. and Potter D. (2004). Low genetic diversity among Frankia spp. strains nodulating sympatric populations of actinorhizal species of Rosaceae, Ceanothus (Rhamnaceae) and Datisca glomerata (Datiscaceae) west of the Sierra Nevada (California). Canad. J. Microbiol. 50: 989–1000
Wallaart R. A. M. (1980). Distribution of sorbitol in Rosaceae. Phytochemistry 19: 2603–2610
Weeden N. and Lamb R. (1987). Genetics and linkage analysis of 19 isozyme loci in apple. J. Amer. Soc. Hort. Sci. 112: 865–872
Wiens J. J. (2003). Missing data, incomplete taxa and phylogenetic accuracy. Syst. Biol. 52: 528–538
Xia X. and Xie Z. (2001). DAMBE: software package for data analysis in molecular biology and evolution. J. Heredity 92: 371–373
Yao C., Conway W. S. and Sams C. E. (1995). Purification and characterization of a polygalacturonase-inhibiting protein from apple fruit. Phytopathology 85: 1373–1377
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Potter, D., Eriksson, T., Evans, R. et al. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266, 5–43 (2007). https://doi.org/10.1007/s00606-007-0539-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-007-0539-9