Abstract
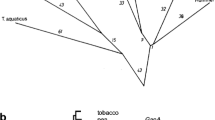
Full-size cDNAs encoding the precursors of chloroplast fructose-1,6-bisphosphatase (FBP), sedoheptulose-1,7-bisphosphatase (SBP), and the small subunit of Rubisco (RbcS) from spinach were cloned. These cDNAs complete the set of homologous probes for all nuclear-encoded enzymes of the Calvin cycle from spinach (Spinacia oleracea L.). FBP enzymes not only of higher plants but also of non-photosynthetic eukaryotes are found to be unexpectedly similar to eubacterial homologues, suggesting a eubacterial origin of these eukaryotic nuclear genes. Chloroplast and cytosolic FBP isoenzymes of higher plants arose through a gene duplication event which occurred early in eukaryotic evolution. Both FBP and SBP of higher plant chloroplasts have acquired substrate specificity, i.e. have undergone functional specialization since their divergence from bifunctional FBP/SBP enzymes of free-living eubacteria.
Similar content being viewed by others
Abbreviations
- FBP:
-
fructose-1,6-bisphosphatase
- SBP:
-
sedoheptulose-1,7-bisphosphatase
- FBA:
-
fructose-1,6-bisphosphate aldolase
References
AbdelalAT, SchlegelHG: Purification and regulatory properties of fructose-1,6-bisphosphatase from Hydrogenomonas eutropha. J Bact 120: 304–310 (1974).
AmachiT, BowienB: Characterization of two fructose bisphosphatase isoenzymes from the hydrogen bacterium Nocardia opaca 1b. J Gen Microbiol 113: 347–356 (1979).
BaalmannE, ScheibeR, CerffR, MartinW: Functional studies of chloroplast glyceraldehyde-3-phosphate dehydrogenase subunits A and B expressed in Escherichia coli: Formation of highly active A4 and B4 homotetramers and evidence that aggregation of the B4 complex is mediated by the B subunit carboxyterminus. Plant Mol Biol 32: 505–513 (1996).
BertschU, SchlicherTB, SchröderI, SollJ: Sequence of mature phosphoglycerate kinase from spinach chloroplasts. Plant Physiol 103: 1449–1450 (1993).
BrinkmannH, CerffR, SalomonM, SollJ: Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol 13: 81–94 (1989).
BrinkmannH, MartinW: Higher plant chloroplast and cytosolic 3-phosphoglycerate kinases: a case of endosymbiotic gene replacement. Plant Mol Biol 30: 65–75 (1996).
BrooksK, CriddleRS: Enzymes of the carbon cycle of photosynthesis. I. Isolation and properties of spinach chloroplast aldolase. Arch Biochem Biophys 117: 650–659 (1966).
BrownJR, DoolittleWF: Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA 92: 2441–2445 (1995).
BuchananBB: Role of light in the regulation of chloroplast enzymes. Annu Rev. Plant Physiol 31: 341–374 (1980).
CadetF, MeunierJ-C: pH and kinetic studies of chloroplast sedoheptulose-1,7-bisphosphatase from spinach (Spinacia oleracea). Biochem J 253: 249–254 (1988).
ClarkCG, RogerAJ: Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc Natl Acad Sci USA 92: 6518–6521 (1995).
DeRijkP, Van dePeerY, Van denBrockI, DeWachterR: Evolution according to large ribosomal subunit RNA. J Mol Evol 41: 366–375 (1995).
DeRocherEJ, QuigleyF, MacheR, BohnertHJ: The six genes of the Rubisco small subunit multigene family from Mesembryanthemum crystallinum, a facultative CAM plant. Mol Gen Genet 239: 450–462 (1993).
Doolittle WF: Some aspects of the biology of cells and their possible evolutionary significance. Symp Soc Gen Microbiol, in press (1996).
FelsensteinJ: PHYLIP: Phylogeny Inference Package (version 3.2). Cladistics 5: 164–166 (1989).
FlechnerA, DreßenU, WesthoffP, HenzeK, SchnarrenbergerC, MartinW: Molecular characterization of transketolase (EC 2.2.1.1) from spinach chloroplasts completes cloning of the Calvin cycle. Plant Mol Biol 132: 475–484 (1996).
FraenkelDG, PontremoliS, HoreckerBL: The specific fructose diphosphatase of Escherichia coli: properties and partial purification. Arch Biochem Biophys 114: 4–12 (1966).
GerblingKP, SteupM, LatzkoE: Fructose 1,6-bisphosphatase form B from Synechococcus leopoliensis hydrolyzes both fructose and sedoheptulose bisphosphate. Plant Physiol 80: 716–620 (1986).
GibsonJL, TabitaFR: Localization and mapping of CO2 fixation genes within two gene clusters in Rhodobacter sphaeroides. J Bact 170: 2153–2158 (1988).
GrossW, BayerMG, SchnarrenbergerC, GebhartUB, MaierTL, SchenkHEA. Two distinct aldolases of the class II type in the cyanoplasts and in the cytosol of the alga Cyanophora paradoxa. Plant Physiol 105: 1393–1398 (1994).
HenzeK, BadrA, WetternM, CerffR, MartinW: A nuclear gene of eubacterial origin in Euglena gracilis reflects cryptic endosymbioses during protist evolution. Proc Natl Acad Sci USA 92: 9122–9126 (1995).
HenzeK, SchnarrenbergerC, KellermannJ, MartinW: Chloroplast and cytosolic triosephosphate isomerase from spinach: purification, microsequencing and cDNA sequence of the chloroplast enzyme. Plant Mol Biol 26: 1961–1973 (1994).
LadrorUS, LatshawSP, MarcusF: Spinach cytosolic fructose-1,6-bisphosphatase. Purification, enzyme properties and structural comparisons. Eur J Biochem 189: 89–94 (1990).
MarcusF, HarrischPB: Amino acid sequence of spinach chloroplast fructose-1,6-bisphosphatase. Arch Biochem Biophys 279: 151–7 (1990).
MartinPG: Amino acid sequence of the small subunit of ribulose-1,5-bisphosphate carboxylase from spinach. Aust J Plant Physiol 6: 401–408 (1979).
MartinW: Is something wrong with the tree of life? BioEssays 18: 523–527 (1996).
MartinW, BrinkmannH, SavonnaC, CerffR: Evidence for a chimeric nature of nuclear genomes: eubacterial origin of glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci USA 90: 8692–8696 (1993).
MartinW, HenzeK, KellermannJ, FlechnerA, SchnarrenbergerC: Microsequencing and cDNA cloning of the Calvin cycle/OPPP enzyme ribose-5-phosphate isomerase (EC 5.3.1.6) from spinach chloroplasts. Plant Mol Biol 30: 795–805 (1996).
MartinW, SomervilleCC, Loiseaux-de GoërS: Molecular phylogenies of plastid origins and algal evolution. J Mol Evol 35: 385–403 (1992).
MilanezS, MuralRJ: Cloning and sequencing of cDNA encoding the mature form of phosphoribulokinase from spinach. Gene 66: 55–63 (1988).
MordenCW, DelwicheCF, KuhselM, PalmerJD: Gene phylogenies and the endosymbiotic origin of plastids. BioSystems 28: 75–90 (1992).
MorseD, SaloisP, MarkovicP, HastingsJW: A nuclear-encoded form II RuBisCO in dinoflagellates. Science 268: 1622–1624 (1995).
MüllerM: Energy metabolism of amitochondriate protists, an evolutionary puzzle. In: SchlegelM, HausmannK (eds) Christian Gottfried Ehrenberg-Festschrift anlässlich der 14. Wissenschaftlichen Jahrestagung der Deutschen Gesellschaft fur Protozoologie, 9.–11. März 1995, Delitzsch (Sachsen), pp. 63–76. Leipziger Universitätsverlag, Leipzig (1996).
NowitzkiU, WesthoffP, HenzeK, SchnarrenbergerC, MartinW: Cloning of the amphibolic Calvin cycle/OPPP enzyme D-ribulose-5-phosphate 3-epimerase (EC 5.1.3.1) from spinach chloroplasts: functional and evolutionary aspects. Plant Mol Biol 29: 1279–1291 (1995).
PalmerJD: Rubisco rules fall; gene transfer triumphs. BioEssays 17: 1005–1008 (1995).
Pelzer-ReithB, PengerA, SchnarrenbergerC: Plant aldolase: cDNA and deduced amino-acid sequence of the chloroplast and cytosol enzyme from spinach. Plant Mol Biol 21: 331–340 (1993).
Pelzer-ReithB, WiegandS, SchnarrenbergerC: Plastid class I and cytosol class II aldolase of Euglena gracilis. Plant Physiol 106: 1137–1144 (1994).
RainesCA, LloydJC, LongstaffM, BradleyD, DyerT: Chloroplast fructose-1,6-bisphosphatase: the product of a mosaic gene. Nucl Acids Res 16: 7931–7942 (1988).
RainesCA, LloydJC, WillinghamNM, PottsS, DyerTA: cDNA and gene sequences of wheat chloroplast sedoheptulose-1,7-bisphosphatase reveal homology with fructose-1,6-bisphosphatases. Eur J Biochem 205: 1053–1059 (1992).
SaitouN, NeiM: The neighbor-joining method: a new method for the reconstruction of phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
SchmidtM, SvendsenI, FeierabendJ: Analysis of the primary structure of the chloroplast isozyme of triosephosphate isomerase from rye leaves by protein and cDNA sequencing indicates a eukaryotic origin of its gene. Biochim Biophys Acta 1261: 257–264 (1995).
SchnarrenbergerC, Pelzer-ReithB, YatsukiH, FreundS, JacobshagenS, HoriK: Expression and sequence of the only detectable aldolase in Chlamydomonas reinhardtii. Arch Biochem Biophys 313: 173–178 (1994).
SpringgateCF, StachowCS: Fructose 1,6-diphosphatase from Rhodopseudomonas palustris. I. Purification and properties. Arch Biochem Biophys 152: 1–12 (1972).
vonHeijneG, StepphuhnJ, HerrmannRG: Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 (1989).
WolterF, FritzC, WillmitzerL, SchellJ, SchreierP: rbcS genes in Solanum tuberosum: conservation of transit peptide and exon shuffling during evolution. Proc Natl Acad Sci USA 85: 846–850 (1988).
YooJG, BowienB: Analysis of the cbbF genes from Alcaligenes eutrophus that encode fructose-1,6-/sedoheptulose-1,7-bisphosphatase. Curr Microbiol 31: 55–61 (1995).
ZimmermannG, KelleyGJ, LatzkoE: Efficient purification and molecular properties of spinach chloroplast fructose 1,6-bisphosphatase. Eur J Biochem 70: 361–367 (1976).
ZurawskiG, PerrotB, BottomleyW, WhitfeldPR: The structure of the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase from spinach chloroplast DNA. Nucl Acids Res 9: 3251–3270 (1981).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Martin, W., Mustafa, AZ., Henze, K. et al. Higher-plant chloroplast and cytosolic fructose-1,6-bisphophosphatase isoenzymes: origins via duplication rather than prokaryote-eukaryote divergence. Plant Mol Biol 32, 485–491 (1996). https://doi.org/10.1007/BF00019100
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00019100